Gene level analyses
We ran gene based tests to look for increased burden of damaging rare variation in cases. First, we annotate variants in the dataset as either ‘synonymous’, ‘other-missense’, ‘damaging-missense’, and ‘PTV’. Here, we define a variant as ‘damaging-missense’ if both the polyphen prediction is ‘probably damaging’ and the SIFT prediction is ‘deleterious’. Then, filtering variants to each of these consequence categories, we performed a number of tests to check for presense of enrichment of rare variation (minor allele count (MAC) \(\leq\) 5).
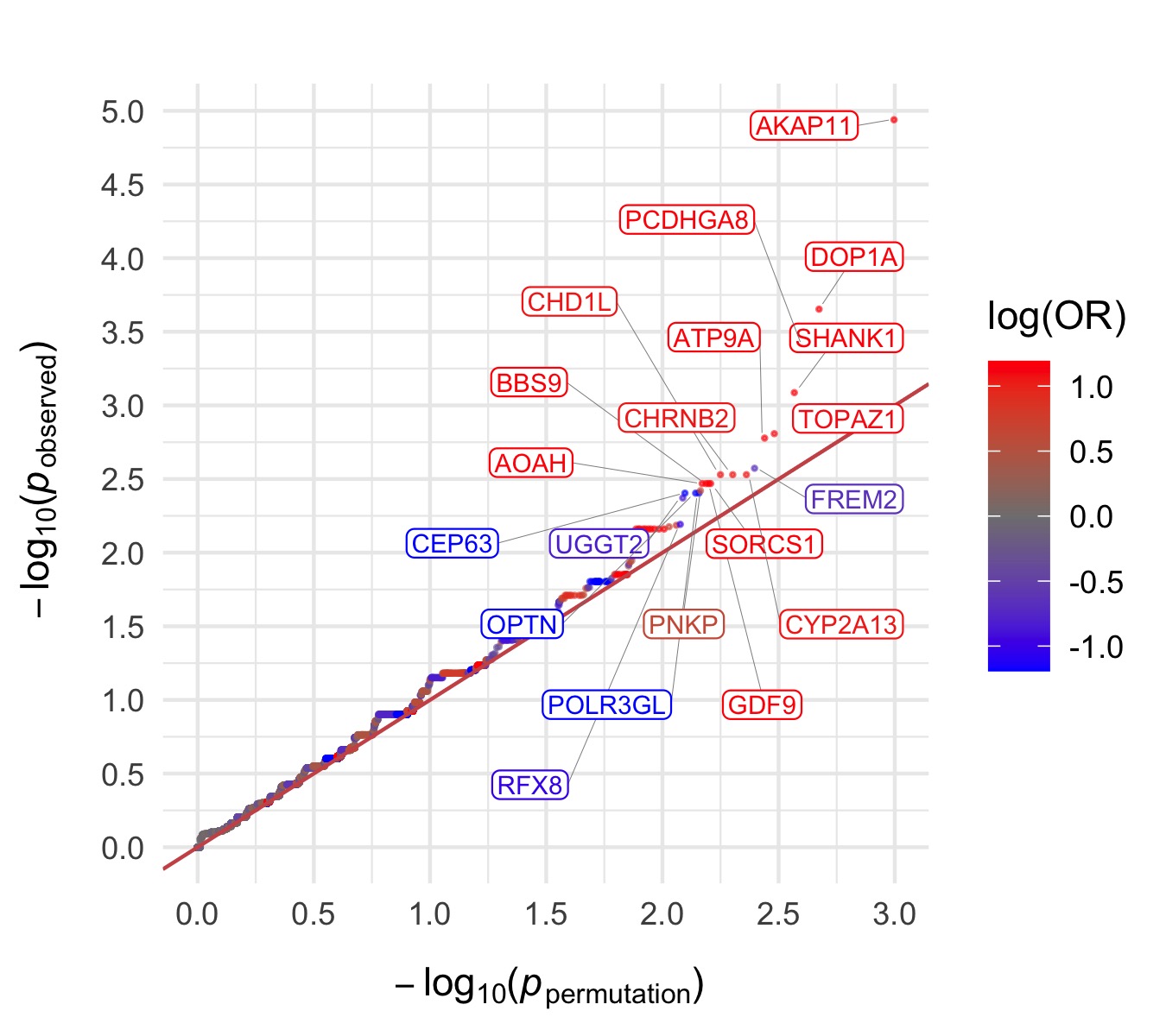
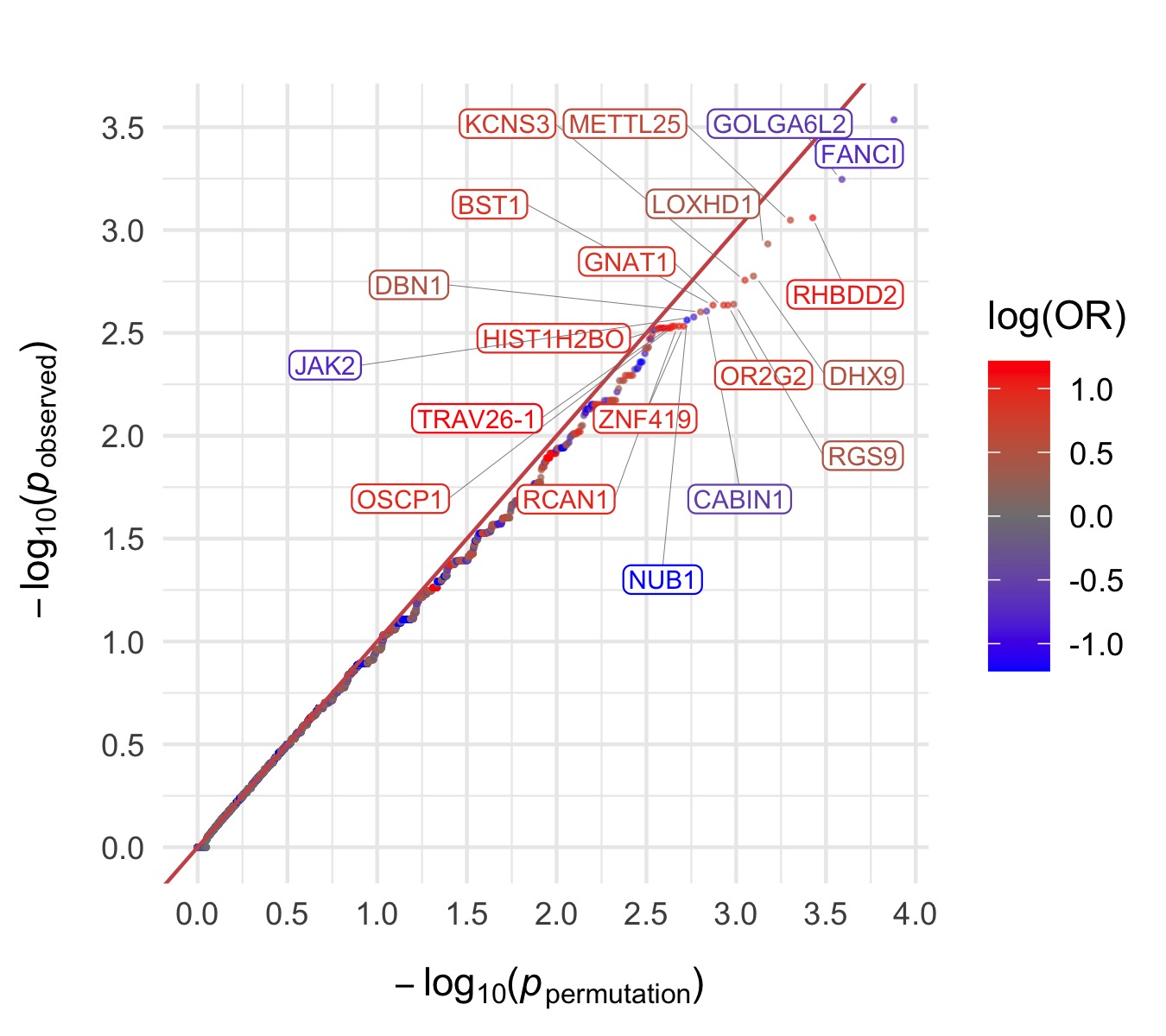
Throughout we show QQ plots next to the gene-based manhattan plots of our results. For each consequence category, we display the results before and after the further restriction of variants not being present in the gnomAD non-psych samples. Non-damaging consequence categories are also included as negative controls. The top 20 genes in each plot by observed \(p\)-value are labelled.
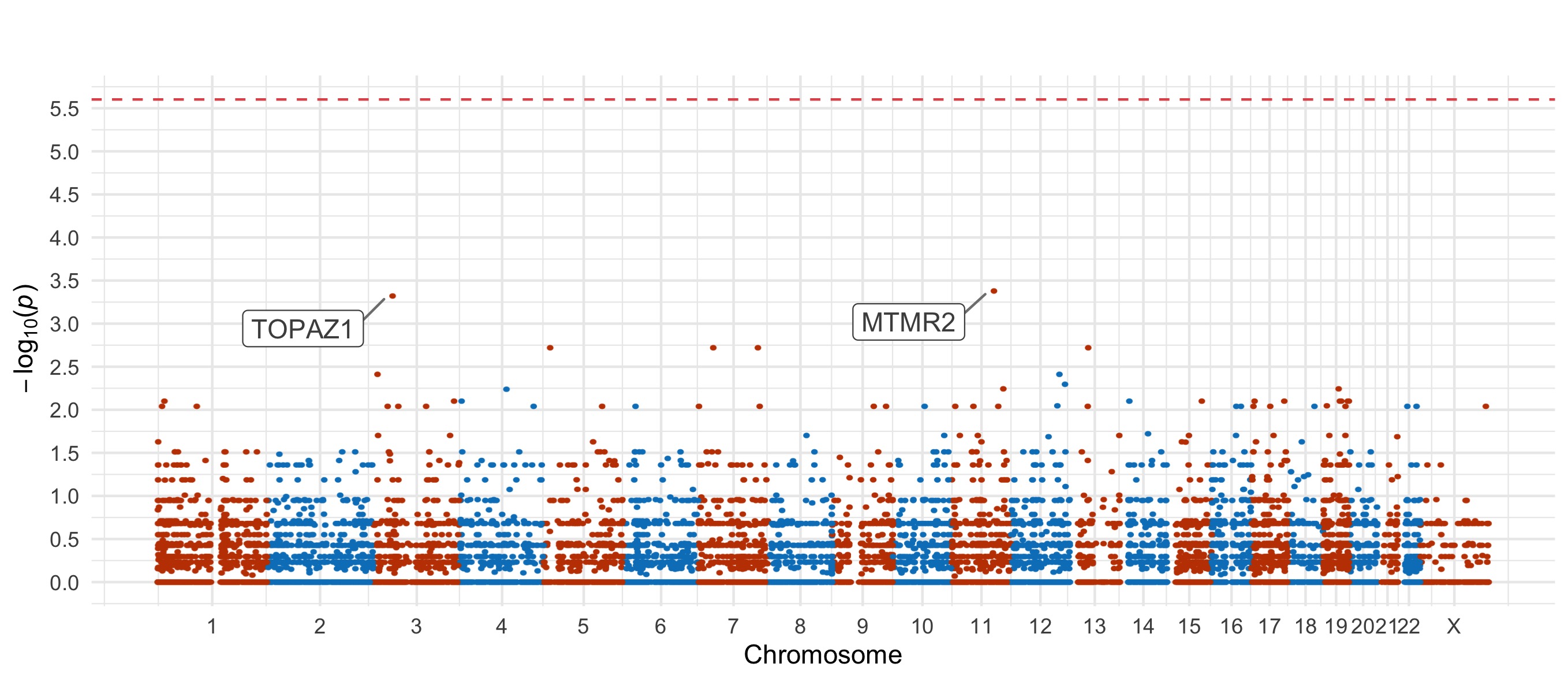
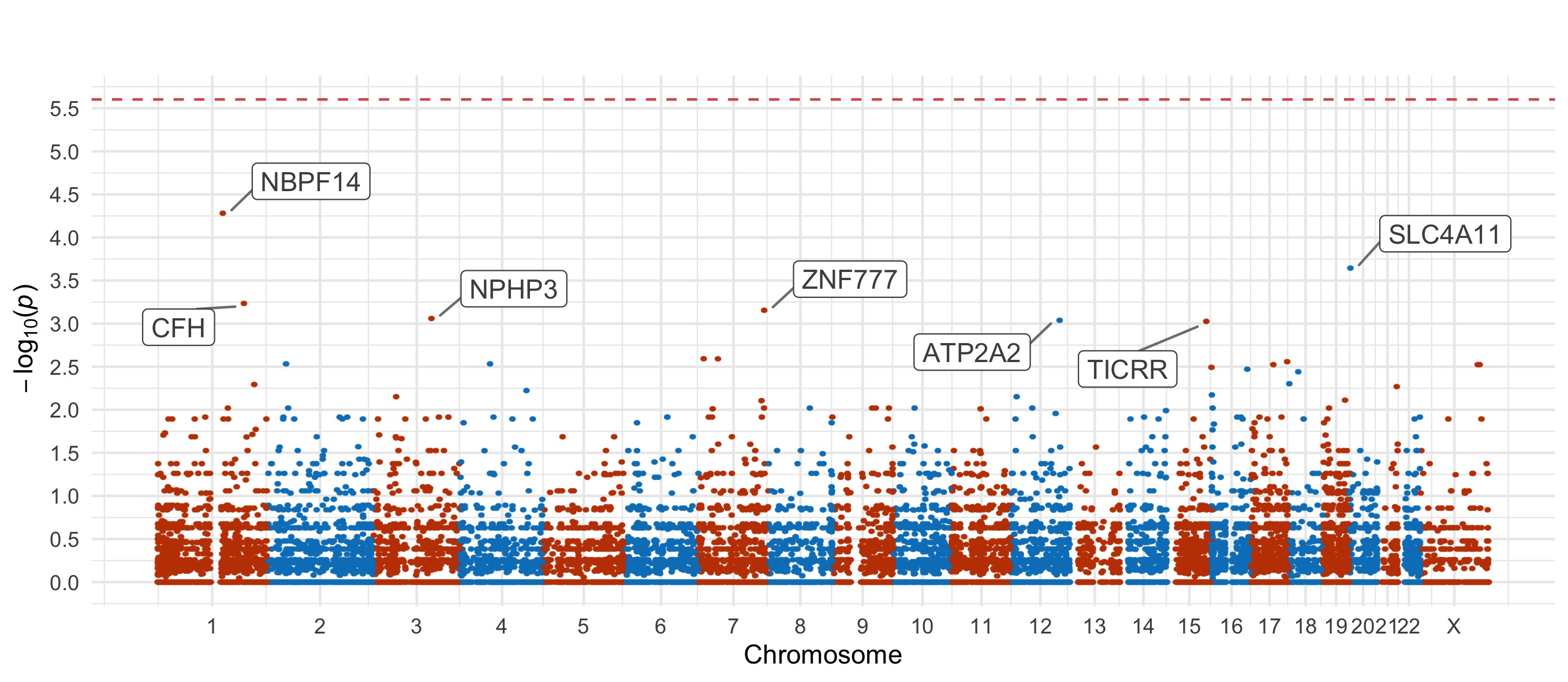
For the damaging missense and PTV tests, we also include Manhattan plots.
Results for the Fisher’s exact tests of MAC \(\leq\) 5 not in the gnomAD non-psych samples can be explored in our browser, take a look at in the ‘Gene Result’ table on the gene specific pages, or here for all the genes. We applied the above tests to bipolar disorder (comprising bipolar disorder without a fine subclassification, bipolar disorder 1, bipolar disorder 2, and bipolar discorder not otherwise specificied), bipolar disorder 1, bipolar disorder 2, and bipolar disorder with and without psychosis. These plots are included in our paper, in Figure 3 and Figures S9-12,14-5.
Fisher’s exact test
We ran a Fisher’s exact test for each gene, grouping together all cohorts. The expected distribution of \(p\)-values is determined via permuation.
Bipolar Disorder
Synonymous

Other missense
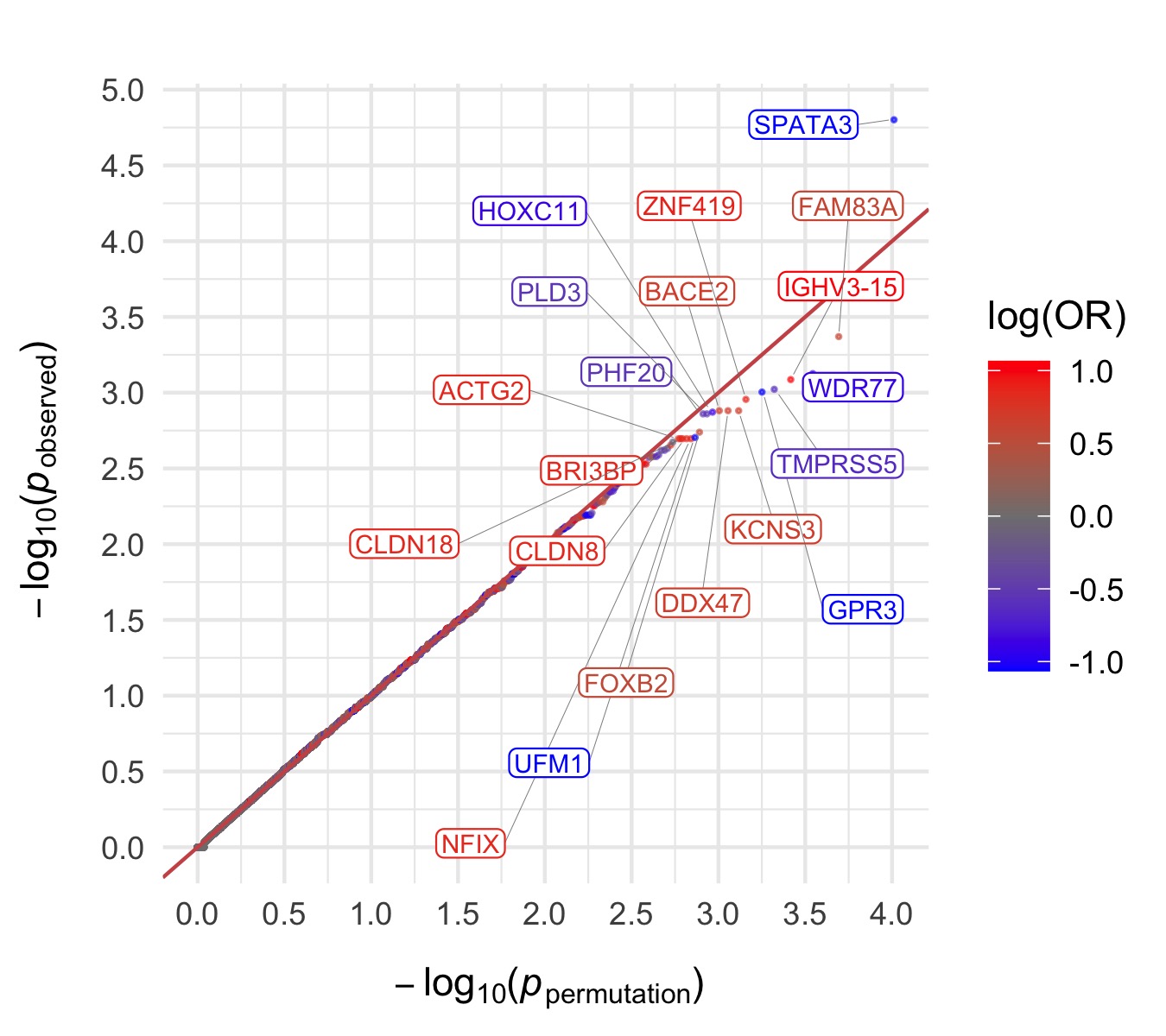
Damaging missense

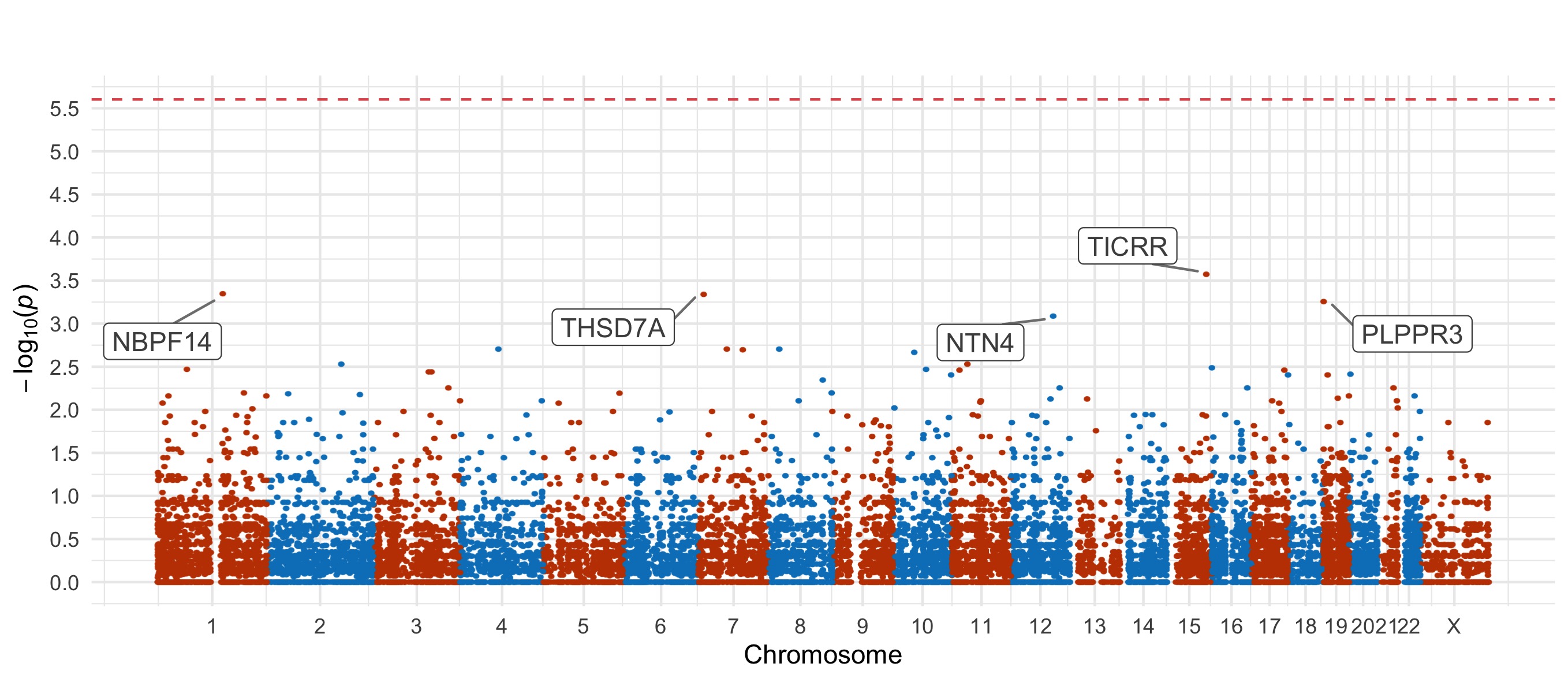
PTV
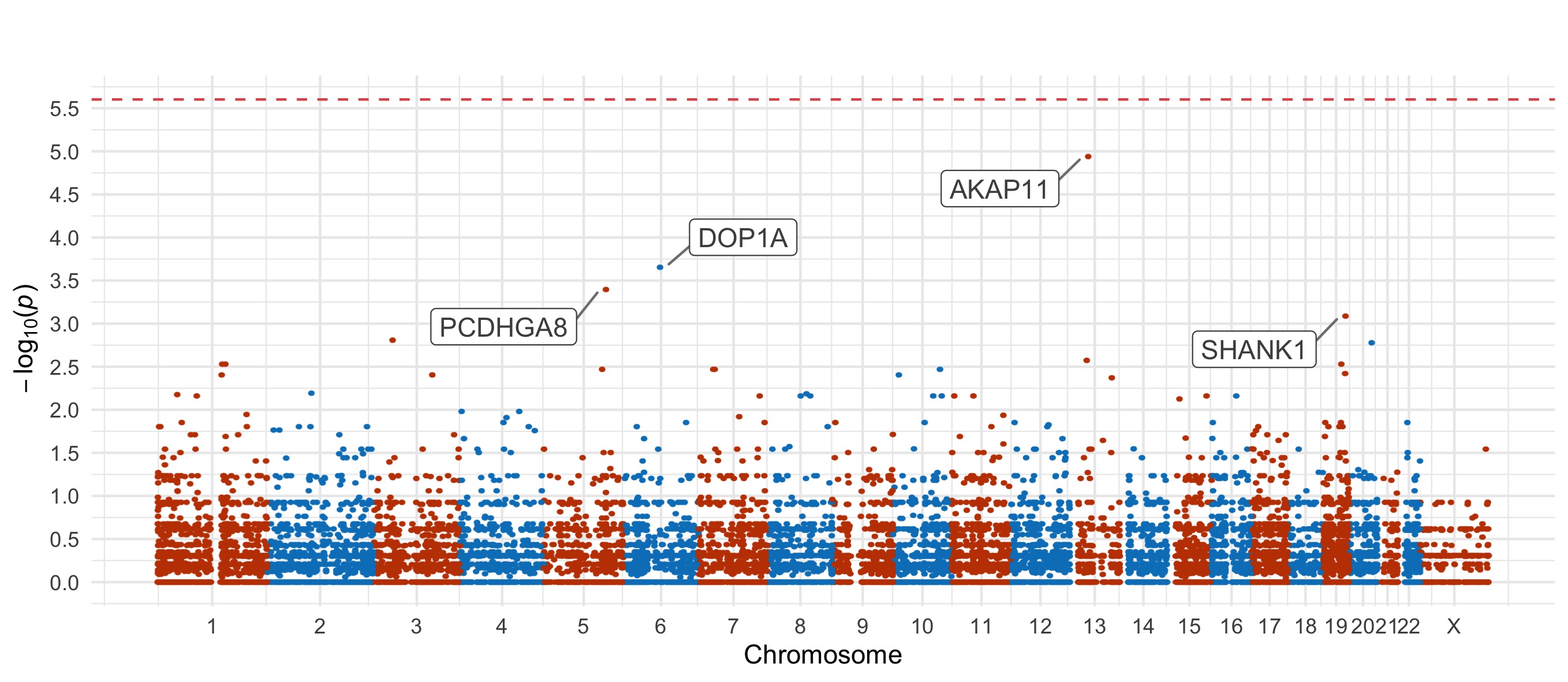
Bipolar Disorder 1
Synonymous

Other missense

Damaging missense


PTV
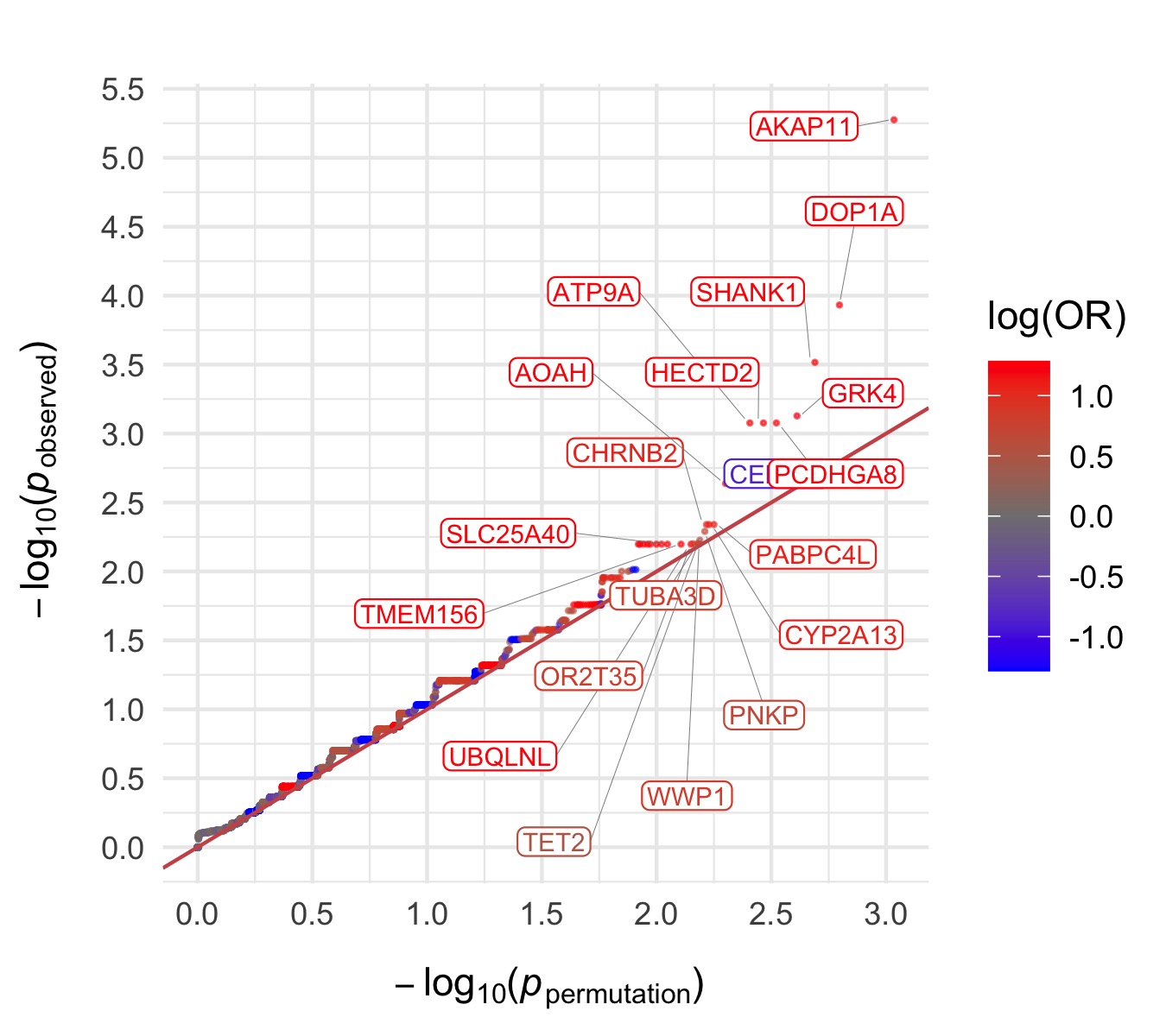
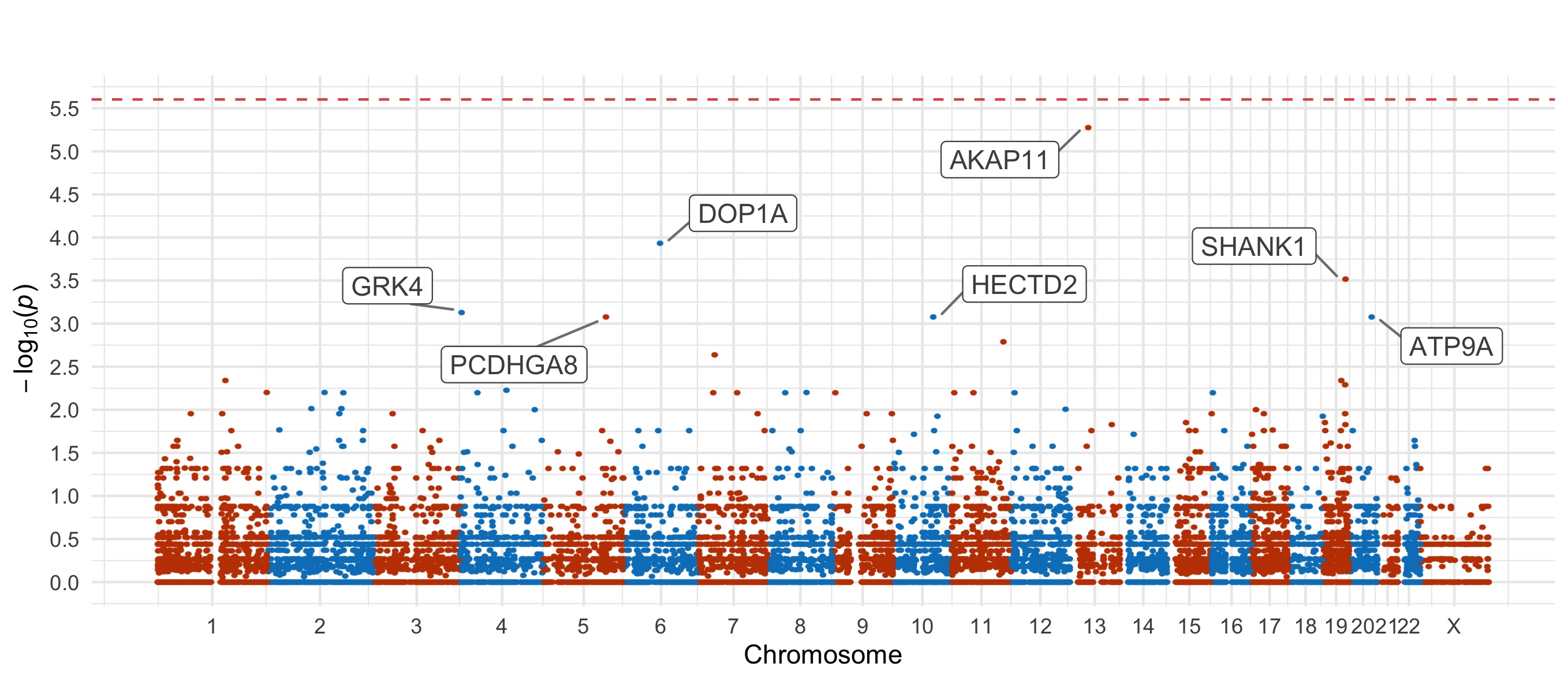
Bipolar Disorder 2
Synonymous

Other missense
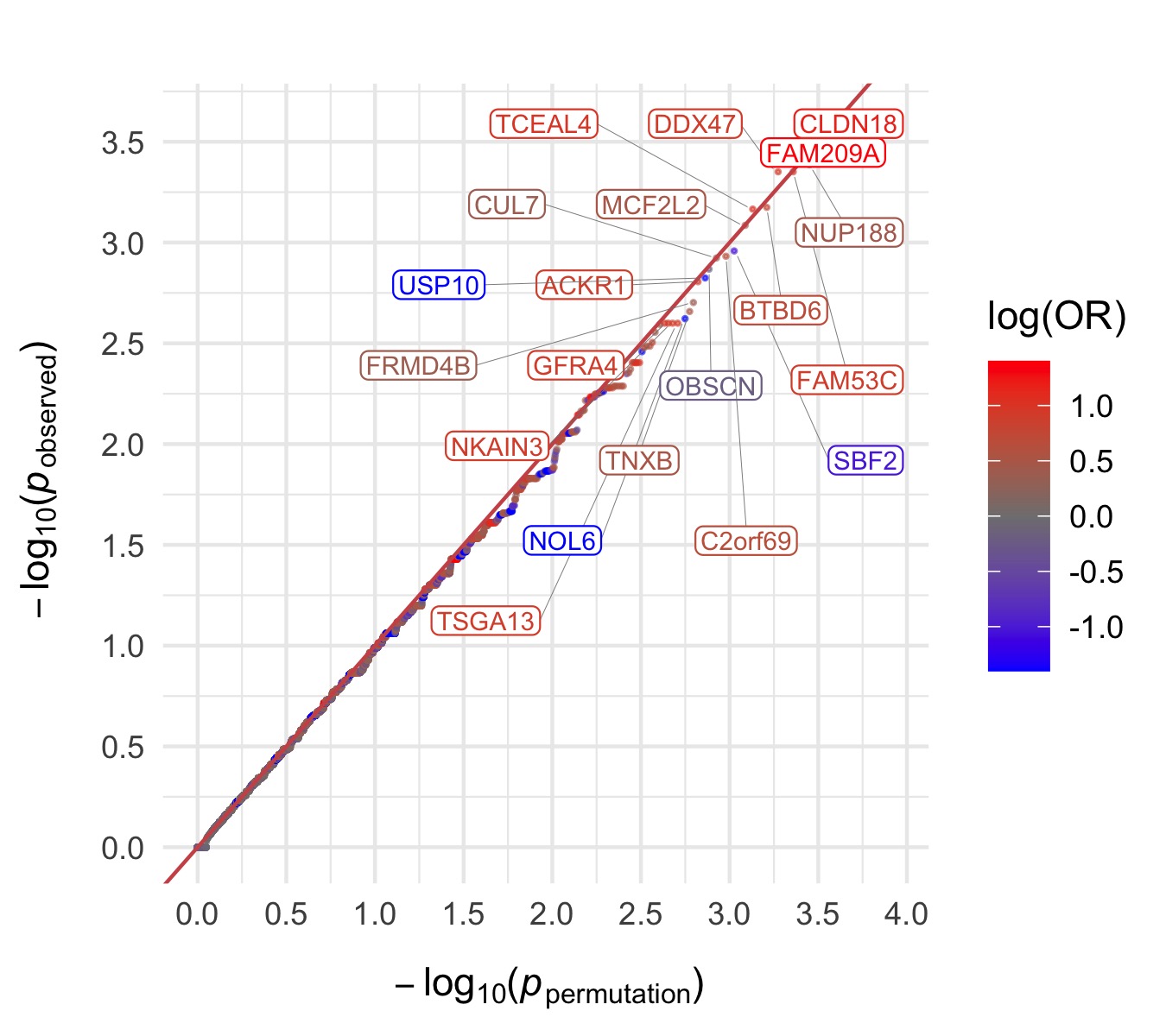
Damaging missense


PTV
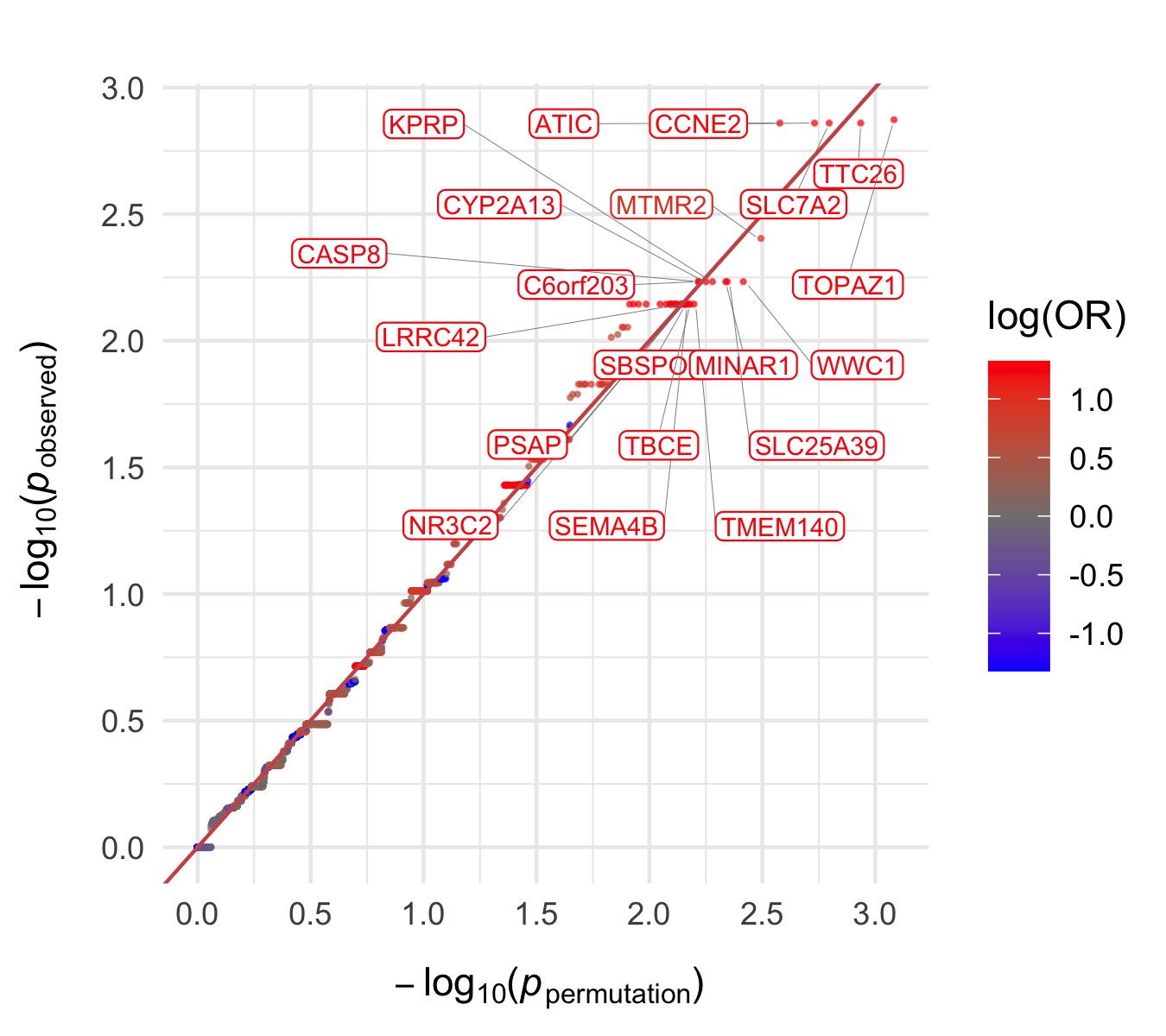
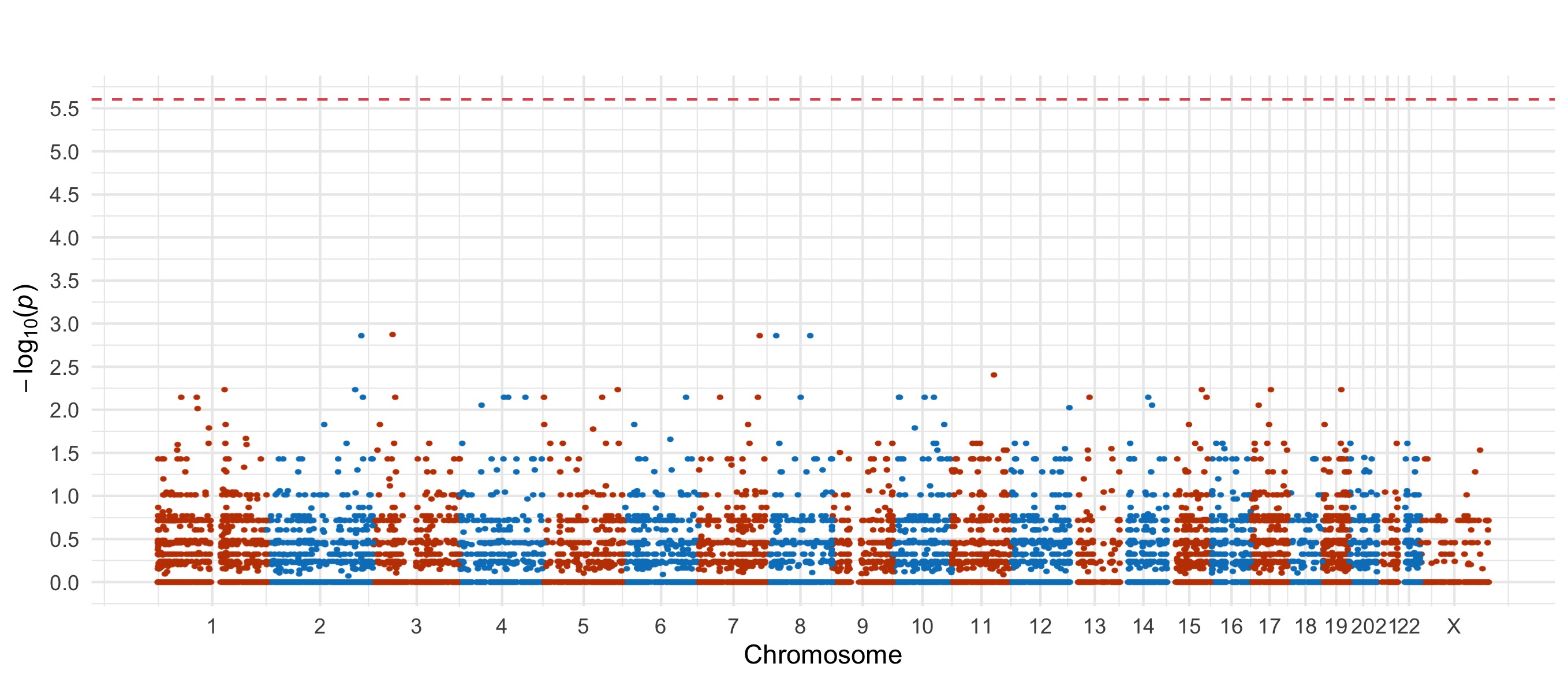
Bipolar Disorder, with Psychosis
Synonymous

Other missense
Damaging missense

PTV
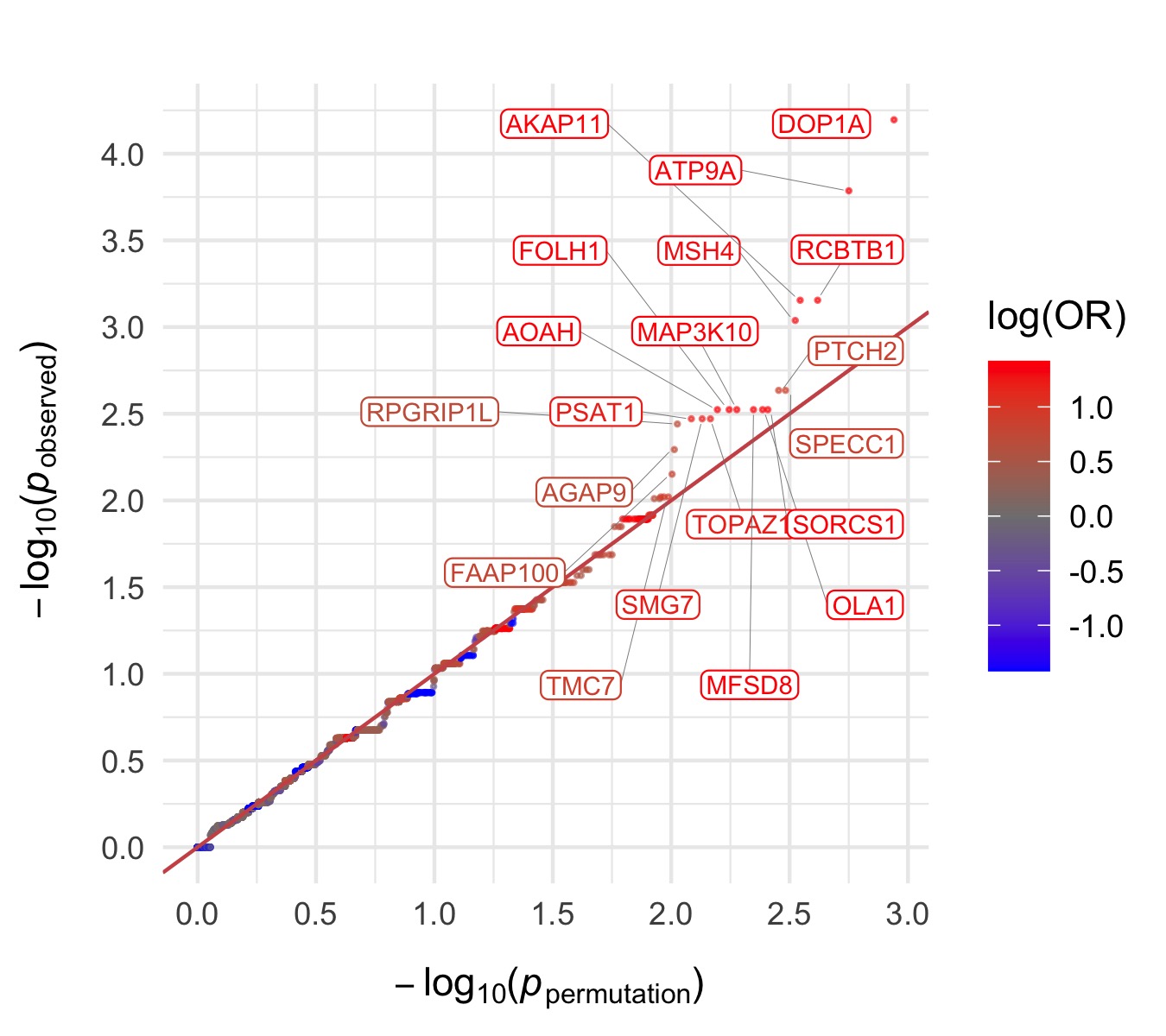
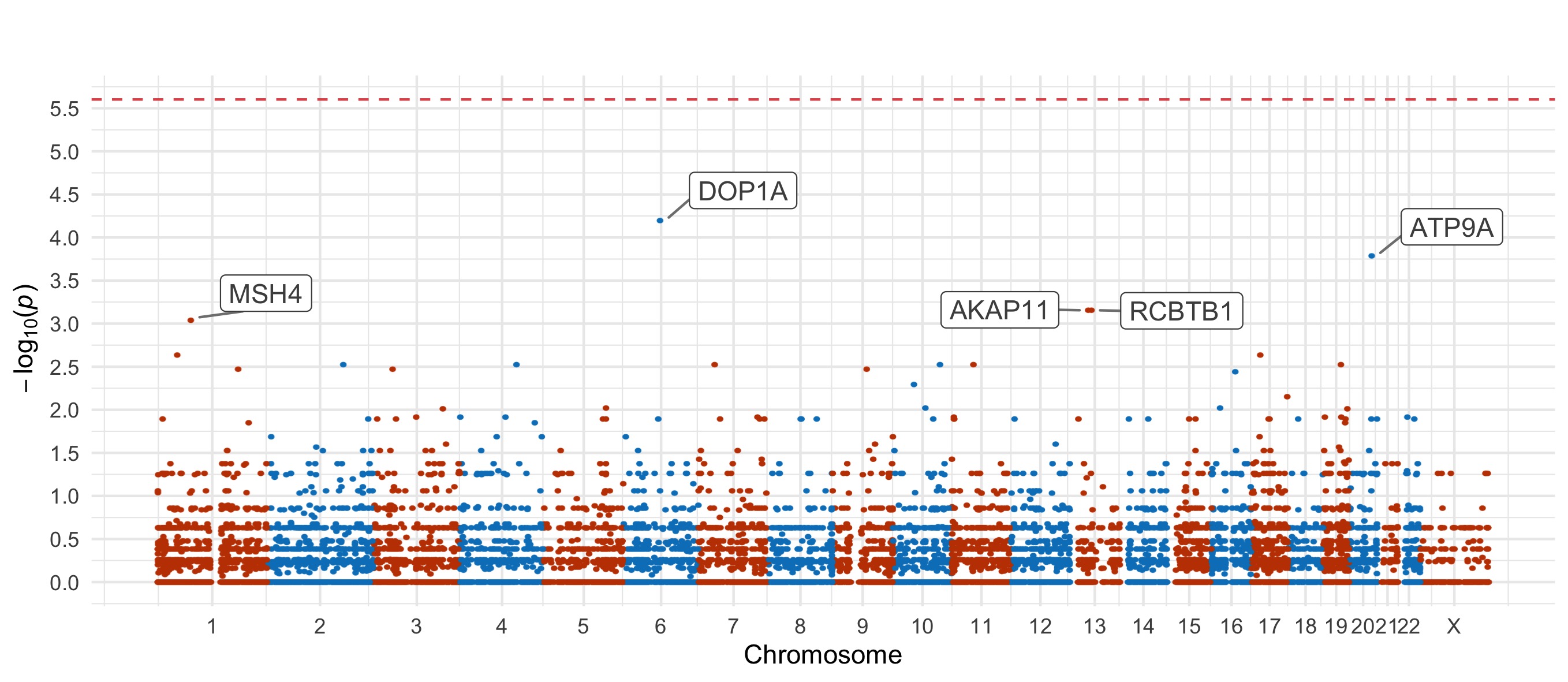
Bipolar Disorder, without Psychosis
Synonymous
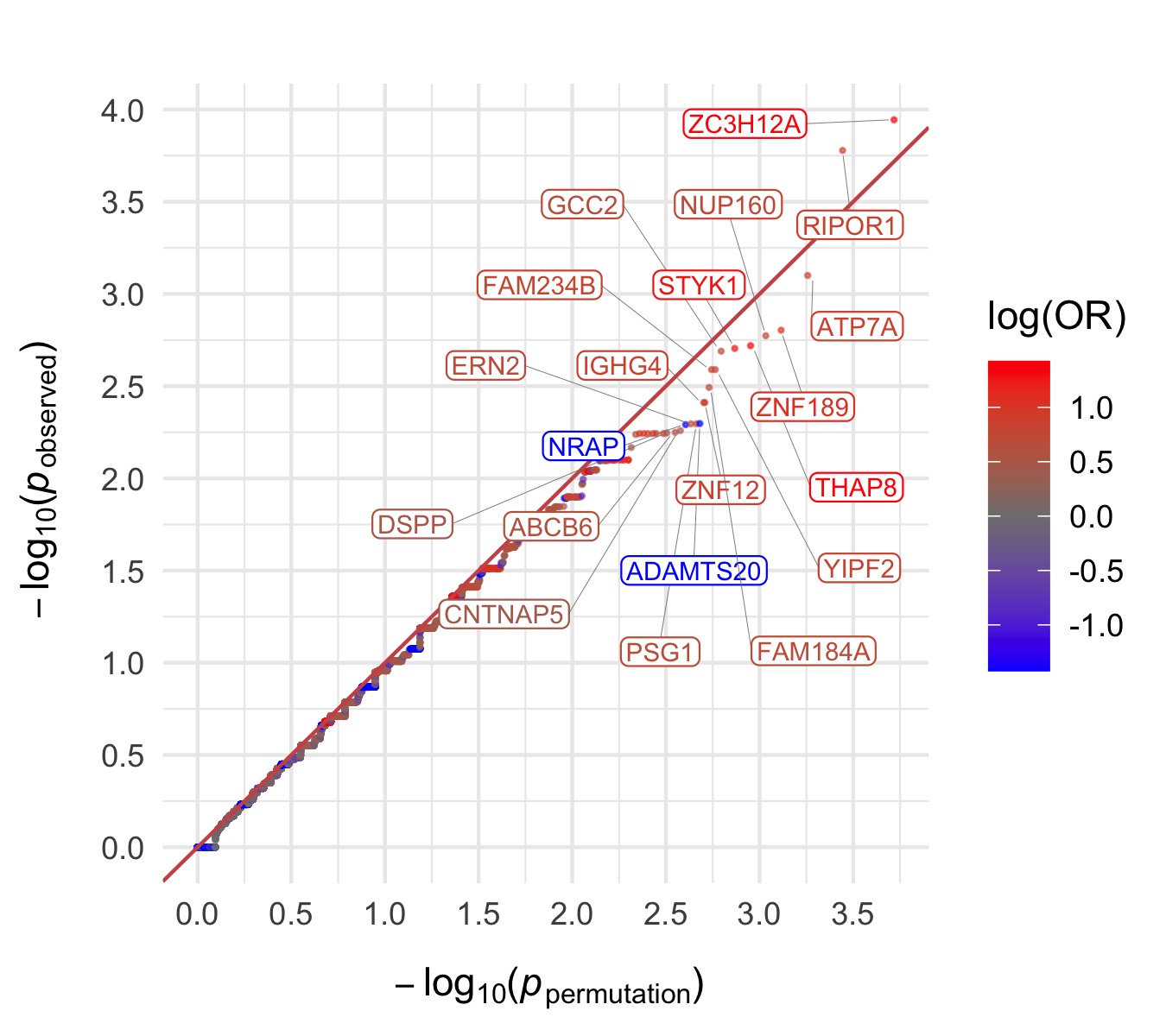
Other missense

Damaging missense

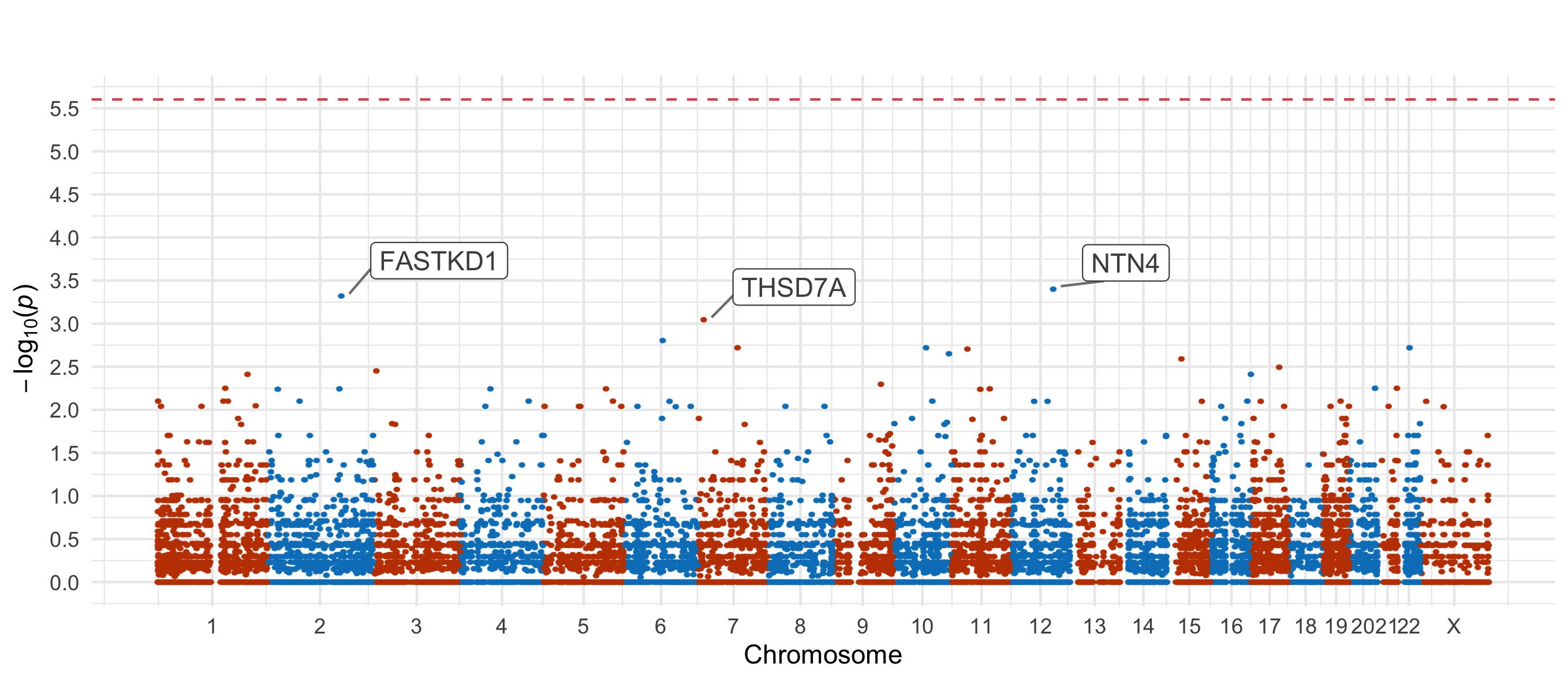
PTV